Simulating BAT Spectra
1. Overview:
This thread describes how to simulate BAT spectra with accurate uncertainties. A special BAT task called 'batphasimerr' is used in conjunction with the standard XSPEC 'fakeit' command to produce the simulated spectrum
Read this thread if you want to: Produce simulated BAT spectra with reasonable error bars.
Last update: 2007-10-06
2. Introduction
Most users are familiar with using XSPEC to produce simulated or "fake" spectra. Such simulated spectra are often needed to support observation proposals, or perhaps to support analysis.
The procedure for simulating BAT spectra is largely the same, but there are some important exceptions. The BAT coded aperture analysis techniques automatically subtract the background from your spectrum, but the level of background does affect the amount of noise the spectrum. This is especially important because BAT is background-dominated for all but the brightest GRBs. XSPEC does not know about any of this, so there are a few extra steps in making a reasonable simulated spectrum.
The steps can be summarized as follows:
Use XSPEC to make a "fake" spectrum as usual, except that...
Answer "NO" to randomization, and set the exposure to 1E9 seconds (infinite exposure time).
Use the 'batphasimerr' task to create the correct uncertainties.
Please note: a new version of WebSpec will be available shortly (Fall 2007) which automates these steps. For simple simulation tasks, WebSpec will almost always be the easiest method.
3. Prerequisites
knowledge of which XSPEC model will be used;
response matrix (see below);
a recent version of HEASoft with batphasimerr (6.3 or later);
a recent version of CALDB.
For the sake of this example, let us make a fake spectrum of the Crab (which has photon spectrum which is approximately a power-law, dN/dE = 10.17 E-2.15), with a simulated exposure of 1000 seconds.
4. Decide on Which Response Matrix To Use
This procedure requires you to use a response matrix. For generic simulations, the most straightforward way to obtain a response matrix is to use the "typical" one stored in CALDB. This matrix represents the response of the BAT to an on-axis source with all detectors enabled (i.e. it is the most optimistic configuration).
If you are not adept at navigating the CALDB directory structure, you can use the following command to retrieve the newest file,
quzcif Swift BAT - - "SPECRESP MATRIX" now now -
which should result in a file name printed to the console.
If you already have an observation which you are processing, you may wish to use the same response matrix.
For the purposes of this example, let's say that the name of the response matrix is, swbresponse20030101v007.rsp.
5. Create the Model Spectrum
For our example, we are simulating a power-law spectrum, which will involve the XSPEC 'powerlaw' model with photon index 2.15 and normalization 10.17 (photons s-1 cm-1 at 1 keV).
In XSPEC, setting up this model will look something like this:
model power law & 2.15 & 10.17
which should result in a display which confirms that the model was set up correctly.
---------------------------------------------------------------------------
Model: powerlaw<1>
Model Fit Model Component Parameter Unit Value
par par comp
1 1 1 powerlaw PhoIndex 2.15000 +/- 0.00000
2 2 1 powerlaw norm 10.1700 +/- 0.00000
---------------------------------------------------------------------------
Of course, you would enter your own spectral model at this stage. Please see the XSPEC home page and documentation for more information about how to use XSPEC.
6. Create the Fake Spectrum
Next, use the XSPEC "fakeit none" command to make a simulated spectrum. This command will prompt you for several inputs.
Run fakeit command.
XSPEC> fakeit none
Enter the name of the response matrix. We are using swbresponse20030101v007.rsp for this example, as described above.
For fake data, file # 1 needs response file: swbresponse20030101v007.rsp
Enter 'none' for the name of the ancillary response file (ARF). There is no need to enter an ARF file since BAT does not use ARFs.
... and ancillary response file: noneEnter "NO" for randomization. The randomization will happen later.
Use randomization in creating fake data? (y) NO
Enter any string for the 'prefix' (it is not used anyway).
Input optional fake file prefix (max 4 chars): dummy
Enter a name for your new fake spectrum. Here we will call it crab.pha and ignore the default.
Fake data filename (...) [/ to use default]: crab.pha
Set the exposure to a large number (1E9 = 1 billion seconds). The correct exposure will be applied later.
Exposure time, correction norm (1, 1): 1E9, 1
The result should be a file called crab.pha which has the correct count rate, but incorrect exposure and uncertainties.
Note that the above series of prompts can be scripted as follows:
fakeit none & swbresponse20030101v007.rsp & none & n & dummy & crab.pha & 1e9
7. Adjust the Spectrum For Background and Exposure
The fake spectrum file produced by XSPEC has the right spectral shape but the incorrect exposure and uncertainties, and no randomization. The BAT task 'batsimphaerr' can insert the correct values into the spectrum, and also randomize the values.
To apply the corrections, issue the following command (either in XSPEC or at the Unix command line):
batphasimerr infile=crab.pha exposure=1000.0 bkgfile=CALDB \ eboundfile=swbresponse20030101v007.rsp pcodefr=1.0
where:
infile is the spectrum file name (the file is modified "in-place");
exposure is the desired exposure in seconds;
bkgfile is the background file (we use CALDB here);
eboundfile is the same as the response matrix we used previously;
pcodefr is the partial coding fraction (1 being the maximum).
After running this command, the crab.pha file should have been corrected.
8. Load Fake Spectrum
At this stage, the corrected file is ready to be used within XSPEC like any other fake spectrum.
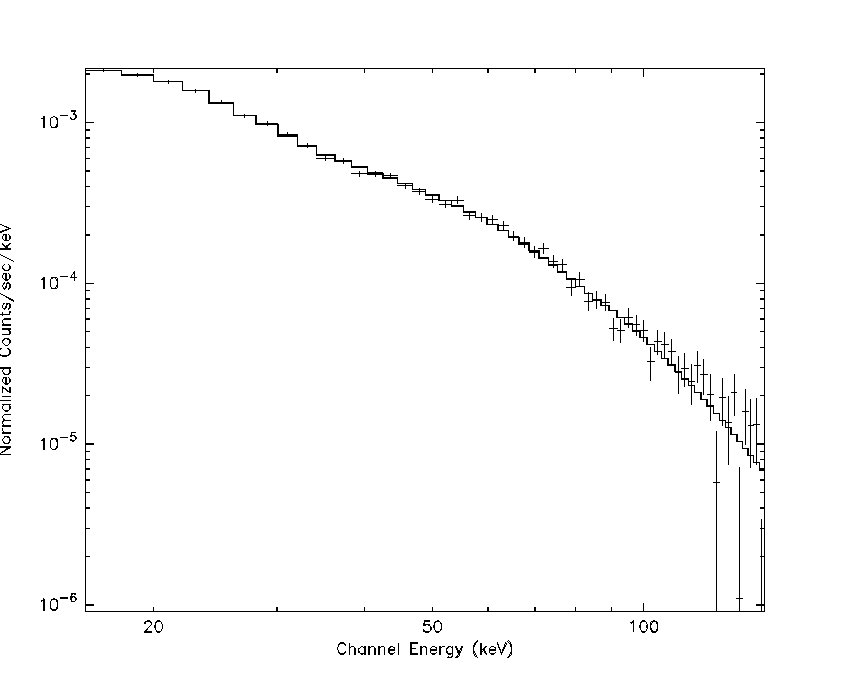
Figure 1. Example simulated Crab spectrum with exposure of 1000 sec. The simulated model is also shown.
Figure shows the result of the simulation described here, as plotted within XSPEC. As noted in the general spectral discussion, you generally should not use GRPPHA for BAT spectral fitting, but for display purposes it is acceptable.
9. Caveats
If you are using the "typical" response matrix, then you should be aware that the simulated spectrum will represent the on-axis fully coded response for all detectors enabled. To simulate off-axis spectra, the simplest approach is to change the partial coding fraction keyword of 'batphasimerr' to the appropriate value.
10. Modifications
2007-10-06 - initial draft.


